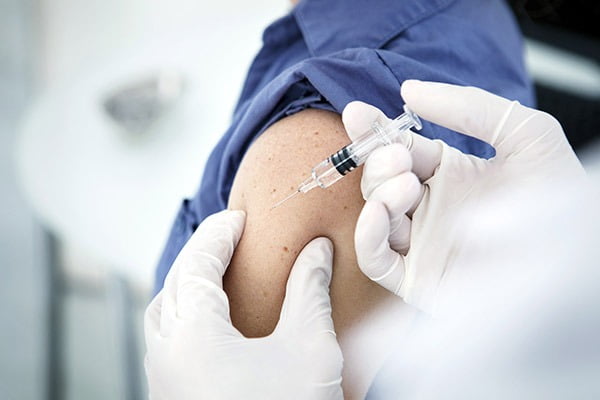
Researchers from UNSW Sydney and the University of Technology Sydney (UTS) have developed a new genomic test for identifying the specific strains of whooping cough. In 2024 all states and territories have reported a rise in whopping cough cases compared to previous years.
The team, led by Professor Ruiting Lan from UNSW and Dr Laurence Luu from UTS, opens in a new window and UNSW, hope to use the assay to pinpoint the strain responsible for the current outbreak in Australia.
“This study paves the way for real-time surveillance of whooping cough strains, overcoming current testing limitations,” says UTS Chancellor’s Research Fellow Dr Laurence Luu, who led the study. “Our results provide important baseline data to understand how whooping cough has changed in Australia and could be used to help guide us through the current outbreak.”
For many years, whooping cough was diagnosed by growing the causative bacteria, before a switch was made over to PCR testing that no longer requires culturing (growing) the live bacteria.
“This change in diagnostic practice means we no longer recover the bacteria for surveillance and can’t monitor what strains are circulating in the community, or whether they are still being targeted by the current vaccine or antibiotics,” says Dr Luu.
To give a clearer insight into the strains causing whooping cough, the team set out to develop a highly sensitive strain test without the need to grow bacteria.
“Usually, to understand what strains are circulating in the community, you need to grow the bacteria, so that you have enough of it to sequence its DNA,” says Dr Luu.
The DNA sequencing on clinical swabs of whooping cough is a cocktail of all sorts of DNA, both from humans and all other bacteria human respiratory tract. “Typically, this makes it really hard to sequence,” says Dr Luu. “But the method we’ve developed, known as mPCR sequencing assay, can directly sequence the residual whooping cough DNA leftover from a PCR test and needs as few as four copies of the bacterial DNA to work effectively. It doesn’t require us to grow the bacteria.”
The team tested their assay on 178 leftover diagnostic DNA samples from across Australia from two previous outbreaks, with the samples collected between 2010-2012 and 2019.
Analysis of the results provided unprecedented detail on the evolution of the whooping cough strains that had been circulating in Australia prior to the pandemic.
One of the key findings revealed that the 2012 outbreak was characterised by up to five different strains, or lineages, of B. pertussis. “We found that the strains that were associated with the big 2008-2012 epidemic had evolved to no longer produce one of the three components that’s targeted by the vaccine.”
By 2019, a single one of these strains had become dominant.
“Having past data is important to understanding how the bacteria may have changed over time, such as whether they are evolving against the vaccines or developing resistance to antibiotics.”
While this latest paper has revealed their assay is effective and sensitive, next steps for Dr Luu and his team involve applying their techniques to the current outbreak.